
ADFilter
Autoencoder filter for publications
This web service is designed to calculate acceptance corrections for any BSM scenario using HEPMC, PROMC, LHE files. It converts truth-level data (from ProMC, HEPMC or LHE files) into identifiable objects such as isolated jets, b-jets, electrons, and muons, which are then processed by publicly available TensorFlow models.
For ATLAS/CMS : ATLAS and CMS can also use HEPMC and LHE files. ATLAS can use PHYS_LIGHT and DAOD inputs. These formats have not been implemented yet.
Supported input files
1) Simple dummy input file
AdFilter supports a number of formats with event records. The best supported is a simple ROOT ntuple which contains 4-momenta of jets, bjets, leptons, photons. The example code that creates an ntuple of this style is given here: dummy.py
wget https://raw.githubusercontent.com/chekanov/ADFilter/main/dummy.py python dummy.py
This creates the file "dummy.root" with 5 objects (1 electron, 1 muon and 1 light jet, 1 photon, 1 b-jet). Use it to upload to the ADFilter. Note that you should also supply a CMS energy and the output files using the arguments of this script. The final output of the loss should log(Loss) = -9.822 (see the histogram "Loss" in the created file "dummy_root_ADFilter.root")
2) ProMC files with truth records
ProMC input files from the HepSim repository. Note such files have only truth-level information with particles, thus the program attempts to create jets, b-jets, isolated leptons and photons. The settings to perform jet reconstruction, b-tagging and isolation are described in the log file of the outputs.
Some files to upload for testing from HepSim database:
(ZprimeZprime 1 TeV), (Charged Higgs), (SSM with 2 TeV Z'), (Composite lepton 2 TeV), (W/Z with filtered lepton), (Zprime bb 2 TeV), (charged Higgs 2 TeV), (ttbar W/Z), (ttbar + N jets), (QCD jets+2l filtered), (QCD jets+l filtered)
3) Delphes ROOT files with fast simulation
One can also use Delphes ROOT files after fast simulations. Unlike ProMC files, Delphes files contain reconstructed jets, b-jets, electrons, muons and photons. You can download Delphes files from the HepSim repository (reconstructed tag rfastXXX, where XXX are some numbers). Here is the direct link Delphes ROOT files.
Since Delphes ROOT files can be rather large, the ADFilter only supports "slimmed" Delphes files for upload. To slim the Delphes ROOT files, you need to have ROOT installation and slimming code from GIT. Then run this command:
git clone https://github.com/wasikatcern/skim_ADFilter.git
cd skim_ADFilter
root -l -q -b 'skim_Delphes.C("original_delphes.root", "skimmed_delphes.root", 13000)'
The number 13000 is the centre-of-mass collision energy (in GeV). We expect that Delphes have the following information (which is default in Delphes):
- Jet reconstructed with anit-kT jets with R=0.4. B-jets are defined as Jet_BTag[j]==1 (Delphes default).
- You should have at least these branches: MissingET, Jet, Electron, Photon and Muon.
- All objects pT (jets, leptons, photons) should have at least 30 GeV cut (not larger!)
- No selection on |Eta|, or it should be at least |Eta| less than 2.4
The output file "skimmed_delphes.root" can be uploaded to the ADfilter. Please use the input file name as unique as possible (not "skimmed_delphes.root" as in this example).
4) LHE files
You can also use LHE files (without parton showers). You need to compress such files using gzip, such that the file extension will be ".lhe.gz". This file will be processed by Pythia8 (for parton showering and hadronization). Then the PROMC file will be produced. Finally, this file will be propagated via the jet reconstruction, exactly as for the original PROMC files.
5) DAOD_PHYS and DAOD_PHYSLIGHT files (ATLAS only)
To upload DAOD_PHYS or DAOD_PHYSLIGHT files (ATLAS data format), you need to slim files to create Ntuples acceptable for ADFilter. Here is the code that does this: DAOD2Ntuple.py.
wget https://raw.githubusercontent.com/chekanov/ADFilter/main/DAOD2Ntuple.py lsetup "asetup Athena,main,latest" python ./DAOD2Ntuple.py --inputlist DAOD_PHYS.37222100._000003.pool.root.1 --outputlist output.root --cmsEnergy 13000 --crossSectionPB 100(here 100 is just a dummy cross section in pb).
Running as a standalone tool on LXPLUS
You can run it on LXPLUS on large files (i.e. Ntuples) like this:/eos/atlas/atlascerngroupdisk/phys-exotics/jdm/lepdijet/adfilter/ADFILTER input_file N /tmp/where "input_file" is the input ROOT (Delphes slimmed), *.lhe.gz or *.promc, "N" is the integer values representing trigger (0 - 7) (check as ADFILTER -h). The last argument is the output directory.
Used Autoencoder
The used autoenoder has about 2 million neurons.
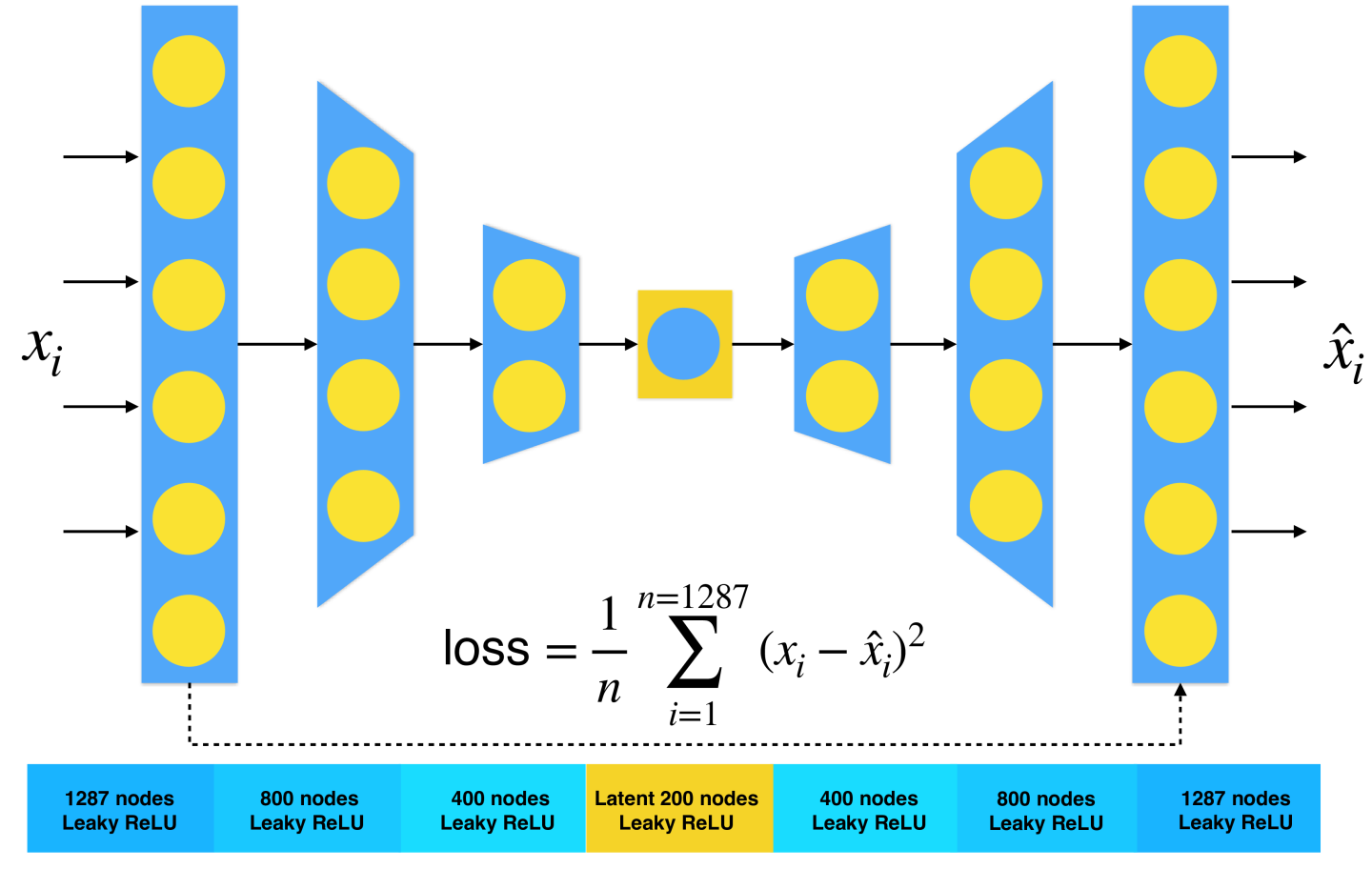
The input is converted 4-momenta into the RMM densities. In the case of different trigger channels, the autoencoder uses "parameterized" approach, i.e. it has additional array indicating which trigger is used.
Output Results:
- ROOT file with reconstructed objects (and RMM) for TensorFlow input
- ROOT file with events, cutflow and invariant masses in the anomaly region.
- Log file with all steps
The ATLAS / JDM anomaly search team